inv_links <- list(
identity = function(eta) eta,
inverse = function(eta) 1/eta,
log = exp)
get_data <- function(inv_link){
x <- pmin(pmax(0, rnorm(n, 2)), 4)
z <- pmin(pmax(0, rnorm(n, 1)), 4)
eta <- 1 + x + 2*z
data.frame(
x=x,
z=z,
eta=eta,
mu=inv_link(eta),
y=rgamma(n, shape=1, scale=inv_link(eta))
)
}
is.in.confint <- function(x, I) (x >I[1]) && (x< I[2])Simulate a glm with exponential distribution such that
\[\operatorname{y} \sim \operatorname{exp}(\operatorname{h}(\alpha + 1\operatorname{x} + 2\operatorname{z}))\]
with the \(h\) being the inverse of the link functions:
- identity
- inverse
- log
Setup
Simulation
For each link function: 1. Plot an example (green line is the true expectation and red line is a smoothing spline) 2. Simulate coverage of true parameter for x (should be 95%)
n <- 100 # sample size
nsim <- 500 # number of simulations to test the confidence interval
set.seed(123)
for (i in seq_along(inv_links)){
D <- get_data(inv_links[[i]])
plot(y ~ mu, main=names(inv_links)[i], data=D)
# Test with SmoothingSplines if indeed: E(y|mu) = mu
with(D, lines(smooth.spline(mu, y), col='red'))
abline(0,1, col='green')
# see if the conf-interval for beta_x is valid. i.e.:
# repeat it nsim times and see if 1 is in our confidence interval
R <- mcreplicate::mc_replicate(nsim,{
try({
D <- get_data(inv_links[[i]])
s <- summary(f <- glm(
y ~ x + z,
data = D,
family = Gamma(link = names(inv_links)[i]),
mustart = mu)); s
suppressMessages(is.in.confint(1, confint(f, "x")))
}, silent = TRUE)
})
coverage <- mean(as.logical(R), na.rm=TRUE)
cat(sprintf("Coverage for %s: %f\n", names(inv_links)[i], coverage))
}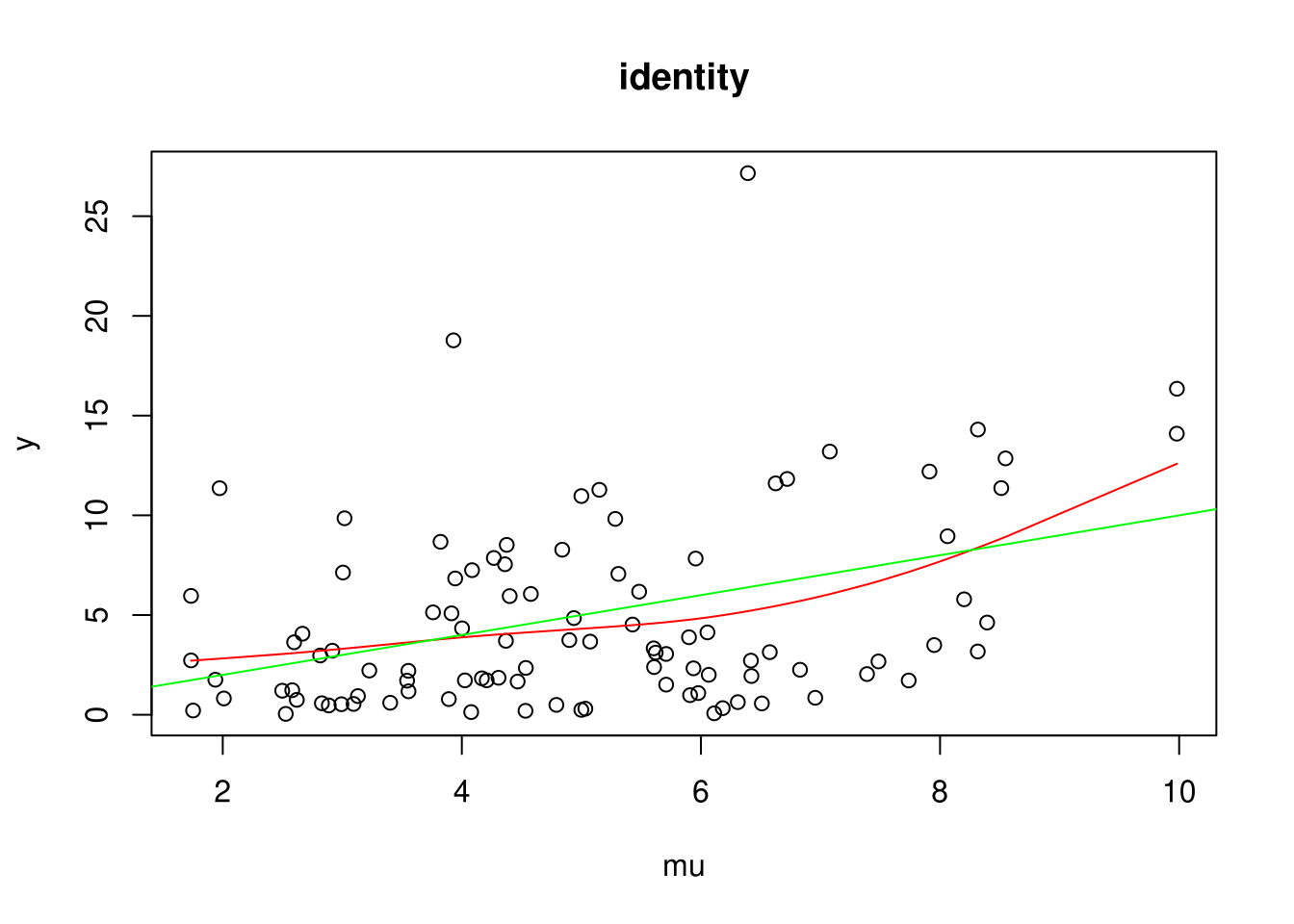
Coverage for identity: 0.940299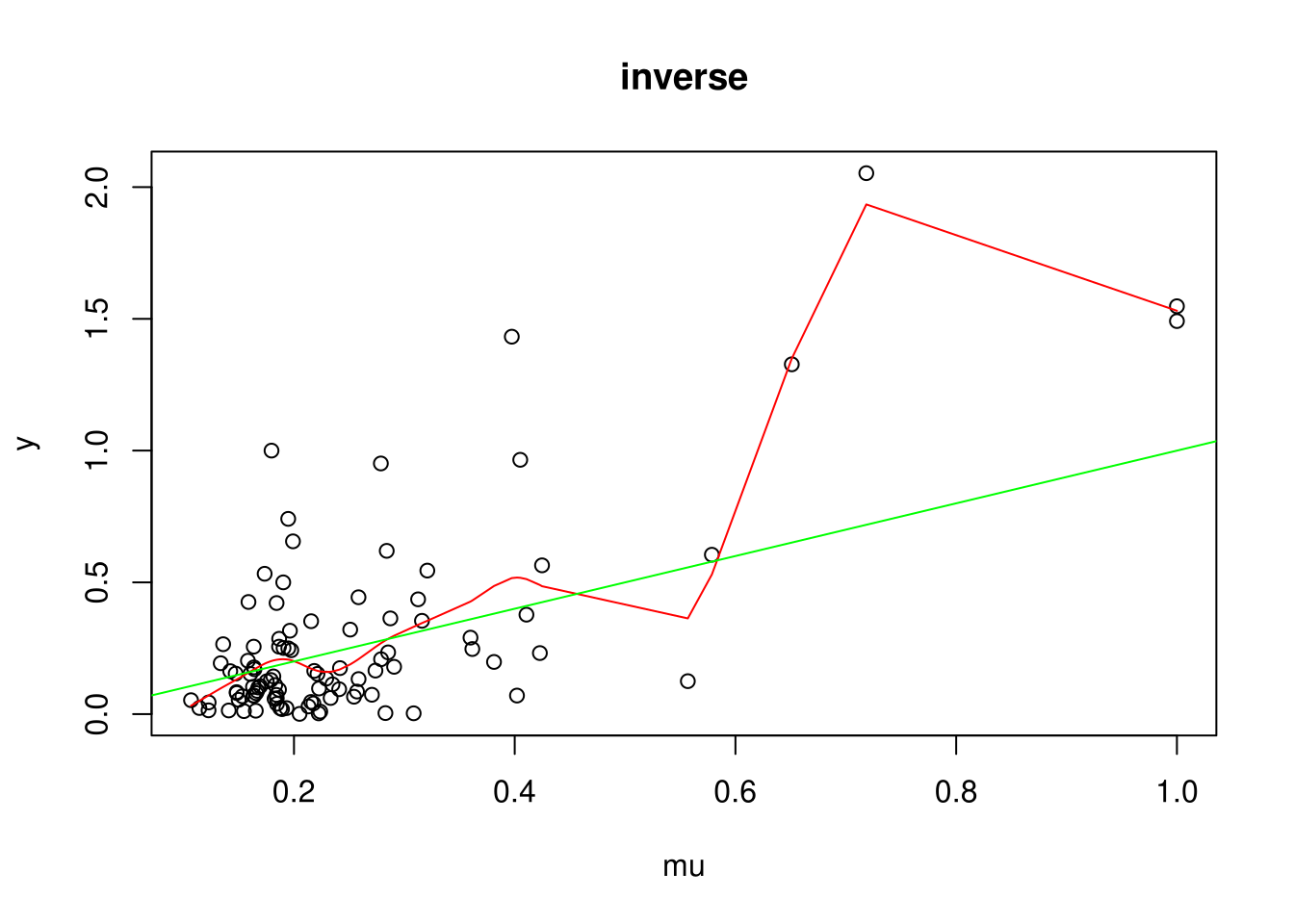
Coverage for inverse: 0.946058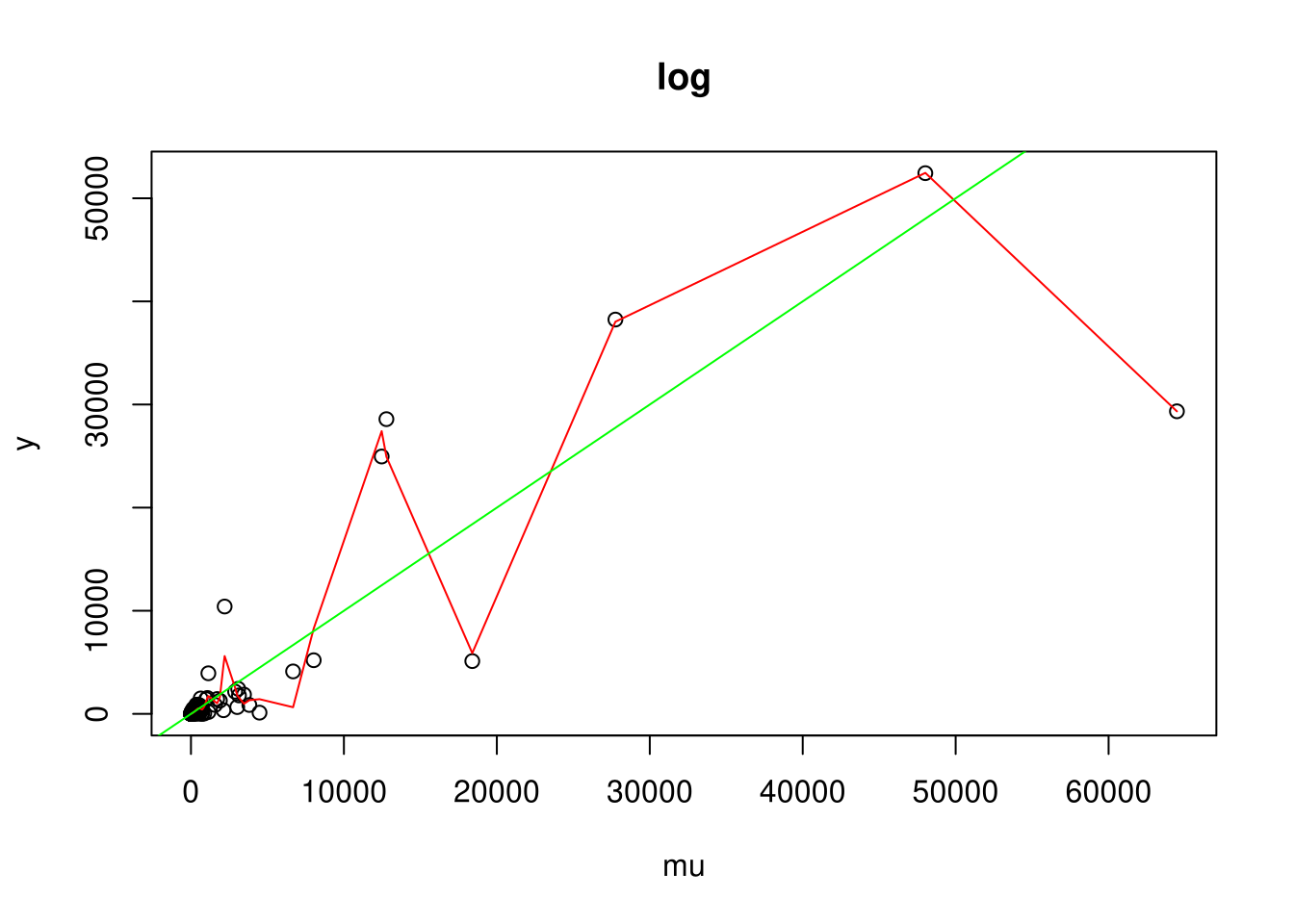
Coverage for log: 0.956000Results
For all link functions, the coverage is about 95%