nsim <- 200
library(coin)
# functions
n <- 40 # per group
laws <- list(
norm = function(mean=0, sd=1) mean + sd * rnorm(n, 0, 1),
logn = function(mean=0, sd=1) mean + sd * (rlnorm(n, sdlog=1) - 1.65) / 1.95,
exp1 = function(mean=0, sd=1) mean + sd * (rexp(n) - 1),
exp2 = function(mean=0, sd=1) mean + sd * (rexp(n)^2 - 2) / 3.94,
emix = function(mean=0, sd=1) mean + sd * (c(rep(0, n/2), rexp(n/2)) - 0.5) / 0.84
)Define a list of laws (i.e., distributions) with mean and sd.
set.seed(123)
options(digits=3)
get_data_all_laws <- function(){
dat <- lapply(names(laws), function(fname){
f <- laws[[fname]]
data.frame(y=f(), fun=fname)
})
dat <- do.call(rbind, dat)
dat
}
boxplot(y ~ fun, get_data_all_laws(), main= "Laws illustrated")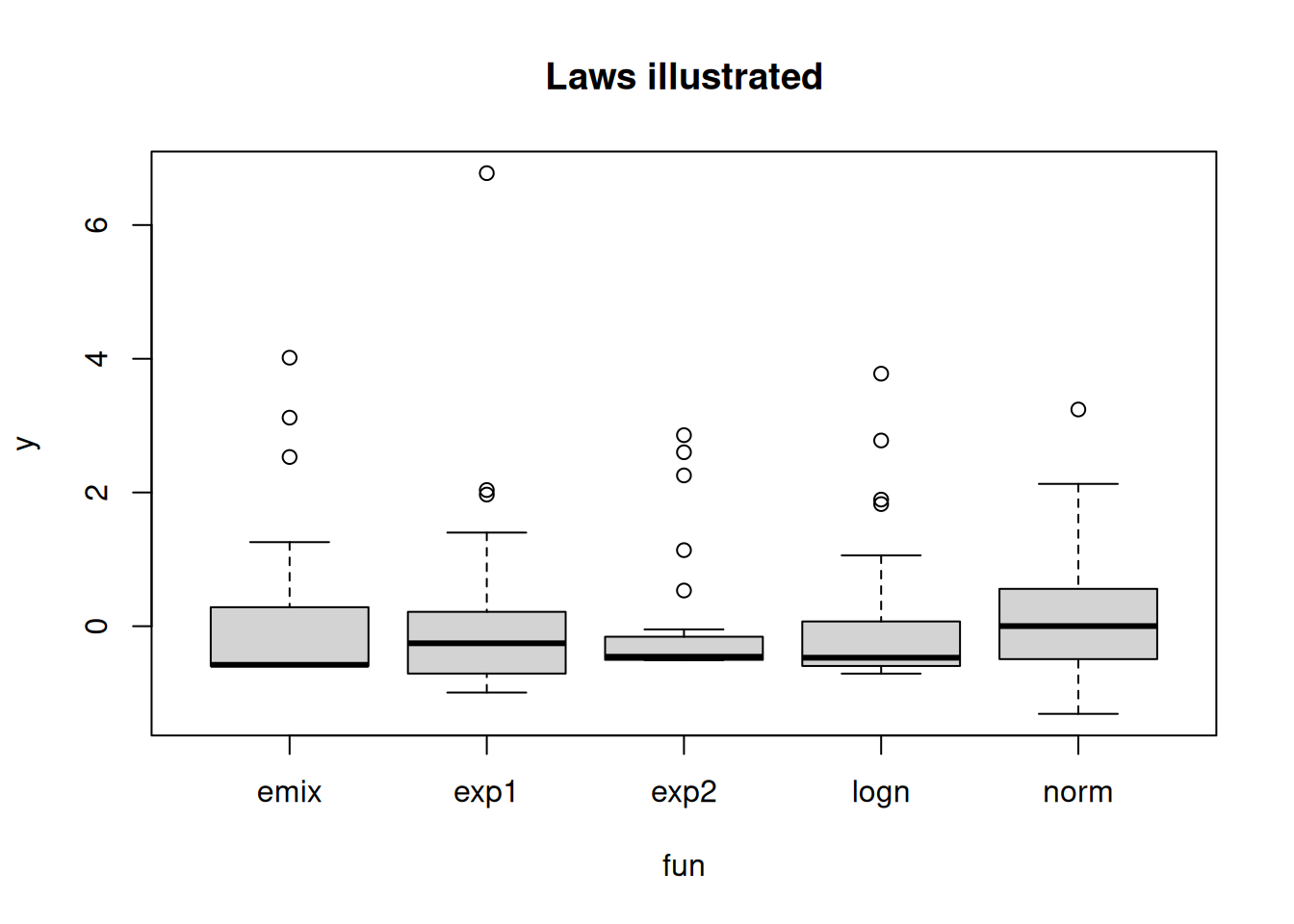
Verify mean and standard deviation
sapply(laws, function(f) mean(replicate(10000, mean(f(0, 1))))) norm logn exp1 exp2 emix
1.55e-03 -2.89e-03 -1.66e-03 -8.53e-05 -1.32e-03 sapply(laws, function(f) mean(replicate(10000, mean(f(1, 2))))) norm logn exp1 exp2 emix
1.003 1.002 0.998 0.994 0.997 sapply(laws, function(f) mean(replicate(10000, sd(f(0, 1))))) norm logn exp1 exp2 emix
0.994 1.008 0.977 0.999 1.000 sapply(laws, function(f) mean(replicate(10000, sd(f(1, 2)))))norm logn exp1 exp2 emix
1.99 2.01 1.96 1.99 2.00 Define the tests used
pval_t <- function(d) t.test(y ~ group,d)$p.value
pval_w <- function(d) coin::pvalue(coin::wilcox_test(y ~ group, d))
pval_m <- function(d) coin::pvalue(coin::median_test(y ~ group, d))# retuns data with f1() for group "A" and f2(mean2, sd2) for group "B"
get_data <- function(f1, f2, mean2=0, sd2=1){
d <- rbind(
data.frame(
y=f1(),
group="A"
),
data.frame(
y=f2(mean2, sd2),
group="B"
))
d$group <- as.factor(d$group)
d
}
# get power of tests
get_power <- function(f1, f2, nsim=1000, mean2=0, sd2=1) {
data_list <- replicate(nsim, get_data(f1, f2, mean2=mean2, sd2=sd2), simplify = FALSE)
c(t = mean(sapply(data_list, pval_t) < 0.05),
w = mean(sapply(data_list, pval_w) < 0.05),
m = mean(sapply(data_list, pval_m) < 0.05))
}
# get all combinations of functions
fun_comb <- expand.grid(names(laws), names(laws)) |> as.matrix()
rnames <- apply(fun_comb, 1, paste0, collapse="_")
sim <- function(nsim=1000, mean2=0, sd2=1) {
fun_comb_list <- split(fun_comb, row(fun_comb))
coverage <- parallel::mclapply(fun_comb_list, function(f_names){
f1 <- laws[[f_names[1]]]
f2 <- laws[[f_names[2]]]
get_power(f1, f2, nsim=nsim, mean2=mean2, sd2=sd2)
})
coverage <- do.call(rbind, coverage)
rownames(coverage) <- rnames
colnames(coverage) <- paste0(
colnames(coverage), " ", as.character(mean2), " ", as.character(sd2))
coverage |> as.data.frame()
}Simulation
set.seed(4321)
null <- sim(nsim=nsim)set.seed(4321)
s_diff <- sim(nsim=nsim, sd2=2)set.seed(4321)
s_difff <- sim(nsim=nsim, sd2=5)set.seed(4321)
mu_diff <- sim(nsim=nsim, mean2 = 0.1)set.seed(4321)
mu_difff <- sim(nsim=nsim, mean2 = 0.2)results <- cbind(
null,
s_diff,
s_difff,
mu_diff,
mu_difff
)
results * 100 t 0 1 w 0 1 m 0 1 t 0 2 w 0 2 m 0 2 t 0 5 w 0 5 m 0 5 t 0.1 1 w 0.1 1
norm_norm 4.0 3.5 5.5 3.0 4.5 4.0 2.0 6.5 4.0 8.5 7.5
logn_norm 5.0 11.5 28.5 6.0 7.5 17.0 5.0 8.0 12.5 10.5 20.5
exp1_norm 2.5 7.0 17.0 5.0 7.5 13.5 3.0 8.0 8.5 11.0 19.5
exp2_norm 9.0 15.0 47.5 6.5 7.0 26.5 3.0 7.5 15.5 10.0 29.0
emix_norm 2.5 11.0 49.5 3.5 8.0 25.5 5.5 10.5 9.5 8.0 19.5
norm_logn 5.5 13.5 29.5 8.0 42.5 59.5 10.5 75.0 74.5 3.0 4.5
logn_logn 3.0 5.5 4.5 4.0 54.0 32.0 9.0 70.5 63.0 8.0 17.0
exp1_logn 7.0 8.0 4.5 10.0 57.5 33.0 12.0 72.0 62.0 7.0 21.0
exp2_logn 2.5 8.0 7.5 6.5 55.5 36.0 10.0 73.5 66.5 9.5 13.0
emix_logn 2.0 3.0 11.0 7.0 61.0 0.5 8.0 74.5 15.5 1.0 29.5
norm_exp1 3.5 9.0 17.5 8.5 26.0 33.5 10.5 48.5 47.5 6.5 6.0
logn_exp1 3.5 6.0 5.5 6.0 35.5 18.0 6.5 45.0 39.0 6.0 6.0
exp1_exp1 3.5 5.5 5.5 6.0 28.5 13.0 8.5 54.5 46.0 7.5 10.0
m 0.1 1 t 0.2 1 w 0.2 1 m 0.2 1
norm_norm 3.5 15.5 14.0 10.0
logn_norm 44.5 21.5 40.5 63.0
exp1_norm 35.0 14.5 29.5 43.5
exp2_norm 64.0 18.0 43.5 76.5
emix_norm 59.0 13.0 37.5 76.5
norm_logn 12.0 11.0 8.5 9.5
logn_logn 9.0 16.5 55.0 27.0
exp1_logn 5.0 12.5 36.0 12.5
exp2_logn 21.5 19.5 47.0 56.5
emix_logn 32.5 14.0 93.5 58.5
norm_exp1 10.5 13.0 4.5 2.0
logn_exp1 9.0 12.0 18.5 16.5
exp1_exp1 8.5 14.0 35.5 17.0
[ reached 'max' / getOption("max.print") -- omitted 12 rows ]Confidence intervals of ratios
prop <- function(ratio, nsim){
confint_ <- prop.test(round(ratio*nsim), nsim)$conf.int[1:2]
names(confint_) <- c("lower", "upper")
c(
ratio= ratio,
confint_
)}
sapply(c(0:4/40, 3:10/20), prop, nsim) |> as.data.frame() * 100 V1 V2 V3 V4 V5 V6 V7 V8 V9 V10 V11 V12 V13
ratio 0.00 2.500 5.00 7.50 10.00 15.0 20.0 25.0 30.0 35.0 40.0 45.0 50.0
lower 0.00 0.924 2.56 4.41 6.37 10.5 14.8 19.3 23.8 28.5 33.2 38.0 43.1
upper 2.35 6.056 9.27 12.30 15.23 20.9 26.4 31.7 36.9 42.1 47.2 52.2 56.9this gives an idea of the uncertainty of a ratio given 200 simulations
redo analysis but with groupwise equal medians
Define a list of laws (i.e., distributions) with median and sd
# functions
n <- 40 # per group
# mean == to keep notation consistent
laws <- list(
norm = function(mean=0, sd=1) mean + sd * rnorm(n, 0, 1)
,logn = function(mean=0, sd=1) mean + sd * (rlnorm(n, sdlog=1) - 1.02) / 1.95
,exp1 = function(mean=0, sd=1) mean + sd * (rexp(n) - 0.706)
,exp2 = function(mean=0, sd=1) mean + sd * (rexp(n)^2 - 0.523) / 3.94
,emix = function(mean=0, sd=1) mean + sd * (c(rep(0, n/2), rexp(n/2)) -0.025) / 0.84
)verify median and standard deviation
sapply(laws, function(f) mean(replicate(10000, median(f(0, 1))))) norm logn exp1 exp2 emix
-0.000142 -0.000216 0.002144 0.000204 0.000246 sapply(laws, function(f) mean(replicate(10000, median(f(1, 2))))) norm logn exp1 exp2 emix
1.000 1.001 0.998 1.000 0.999 sapply(laws, function(f) mean(replicate(10000, sd(f(0, 1))))) norm logn exp1 exp2 emix
0.996 1.006 0.978 1.010 1.003 sapply(laws, function(f) mean(replicate(10000, sd(f(1, 2)))))norm logn exp1 exp2 emix
1.99 2.00 1.95 1.99 2.01 boxplot(y ~ fun, get_data_all_laws(), main= "Equal Medians (in expectation)")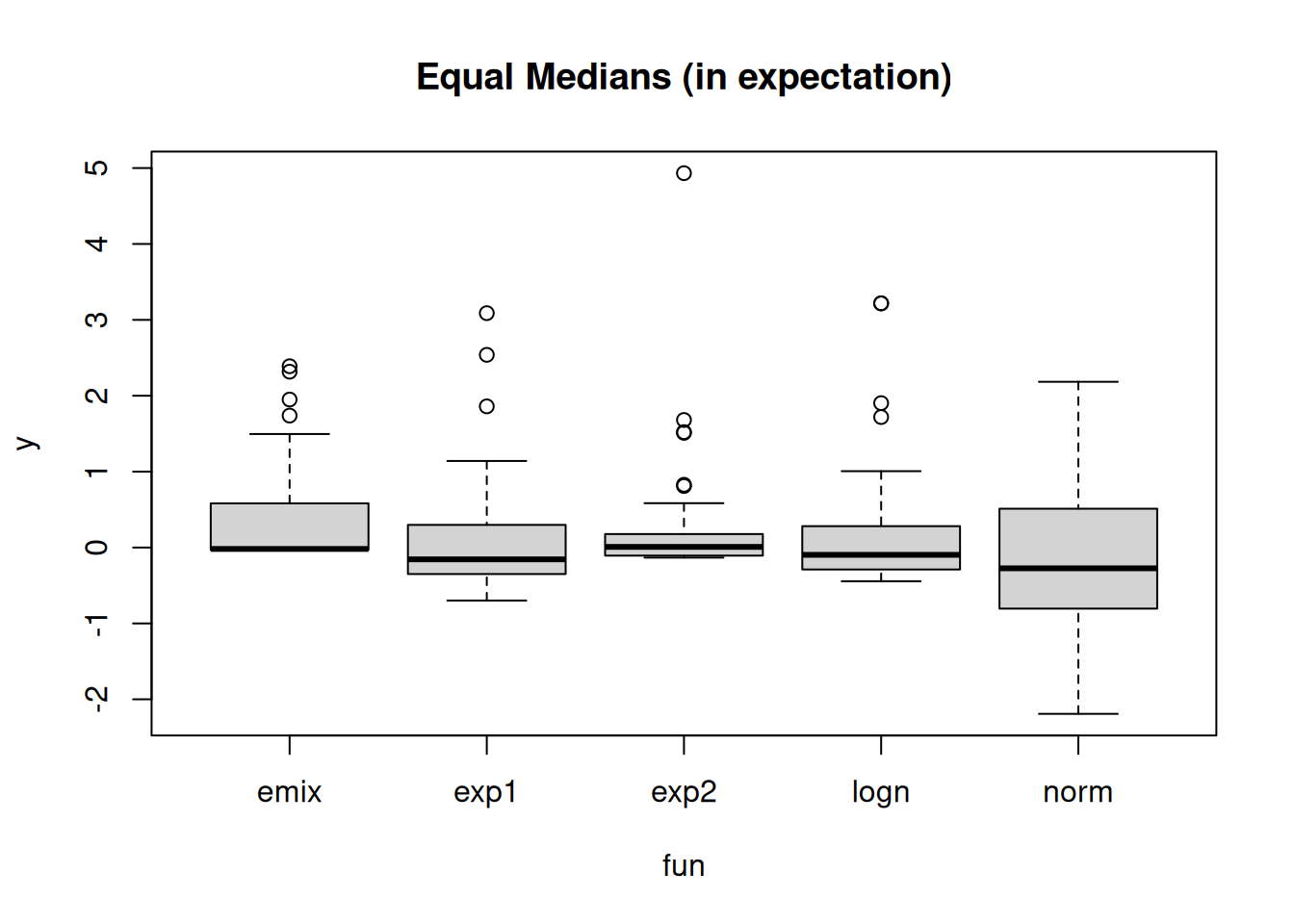
set.seed(4321)
null_median <- sim(nsim=nsim)set.seed(4321)
s_diff_median <- sim(nsim=nsim, sd2=2)set.seed(4321)
s_difff_median <- sim(nsim=nsim, sd2=5)set.seed(4321)
mu_diff_median <- sim(nsim=nsim, mean2 = 0.1)set.seed(4321)
mu_difff_median <- sim(nsim=nsim, mean2 = 0.2)results_median <- cbind(
null_median,
s_diff_median,
s_difff_median,
mu_diff_median,
mu_difff_median
)
results_median * 100 t 0 1 w 0 1 m 0 1 t 0 2 w 0 2 m 0 2 t 0 5 w 0 5 m 0 5 t 0.1 1 w 0.1 1
norm_norm 2.0 2.0 2.5 5.0 6.5 5.5 9.5 11.5 11.5 7.5 7.0
logn_norm 26.0 16.0 5.0 11.5 16.5 11.0 5.0 8.5 9.0 14.0 8.0
exp1_norm 21.5 10.5 6.5 13.0 12.0 8.0 3.0 6.0 8.5 10.5 6.5
exp2_norm 27.5 19.5 7.0 18.0 17.0 10.5 5.5 10.0 13.5 18.5 12.0
emix_norm 75.0 53.0 1.0 30.5 27.5 2.5 12.0 15.5 4.5 55.5 35.5
norm_logn 22.5 11.0 6.0 34.5 13.0 3.5 43.0 9.0 7.5 33.5 27.5
logn_logn 3.5 5.5 3.5 7.0 7.5 6.0 23.5 11.5 10.0 6.5 15.5
exp1_logn 6.0 9.0 6.5 15.5 8.0 5.5 25.5 10.5 8.0 7.5 20.5
exp2_logn 3.0 16.5 5.5 4.5 14.5 6.5 21.5 14.5 11.5 6.0 3.5
emix_logn 26.5 64.5 0.0 4.5 33.0 0.5 17.0 17.0 1.0 9.0 25.0
norm_exp1 27.0 14.5 8.5 39.5 9.0 3.5 46.5 10.0 14.5 44.5 30.0
logn_exp1 4.0 8.0 4.0 13.0 16.5 10.5 21.5 10.5 13.5 8.5 6.0
exp1_exp1 2.5 3.0 5.5 16.0 7.5 8.5 26.5 9.5 7.5 7.0 9.5
m 0.1 1 t 0.2 1 w 0.2 1 m 0.2 1
norm_norm 6.5 12.5 11.5 11.0
logn_norm 6.0 6.5 8.5 11.5
exp1_norm 7.5 7.5 6.5 13.0
exp2_norm 7.5 5.0 5.0 20.5
emix_norm 3.5 34.0 16.5 3.0
norm_logn 10.0 64.5 53.5 17.5
logn_logn 9.5 16.0 51.0 24.5
exp1_logn 9.0 15.5 50.0 24.5
exp2_logn 17.5 13.0 24.0 37.0
emix_logn 1.0 2.5 1.0 3.0
norm_exp1 8.5 57.0 41.5 13.0
logn_exp1 7.5 10.5 13.5 13.5
exp1_exp1 5.0 17.0 30.0 15.5
[ reached 'max' / getOption("max.print") -- omitted 12 rows ]