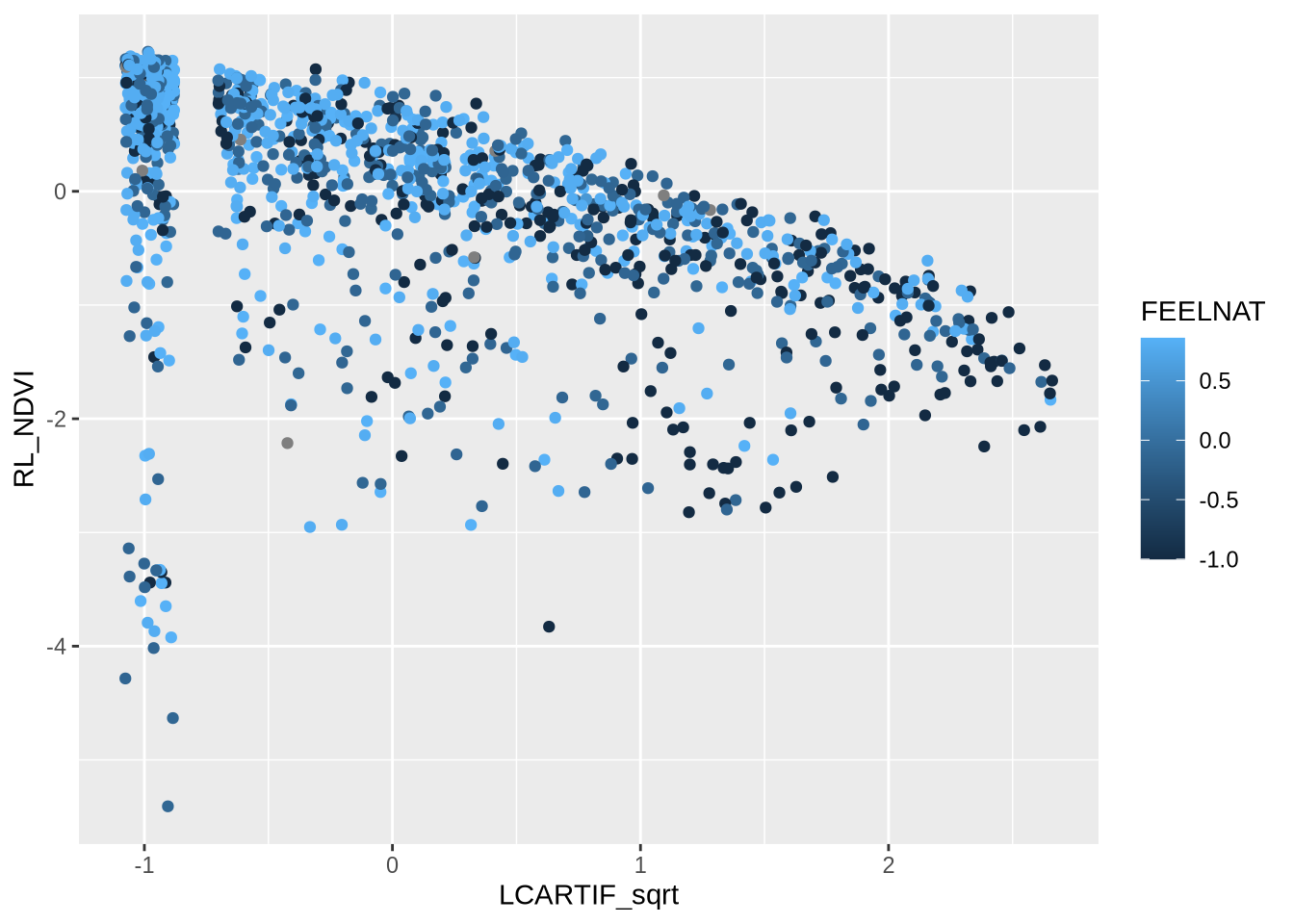
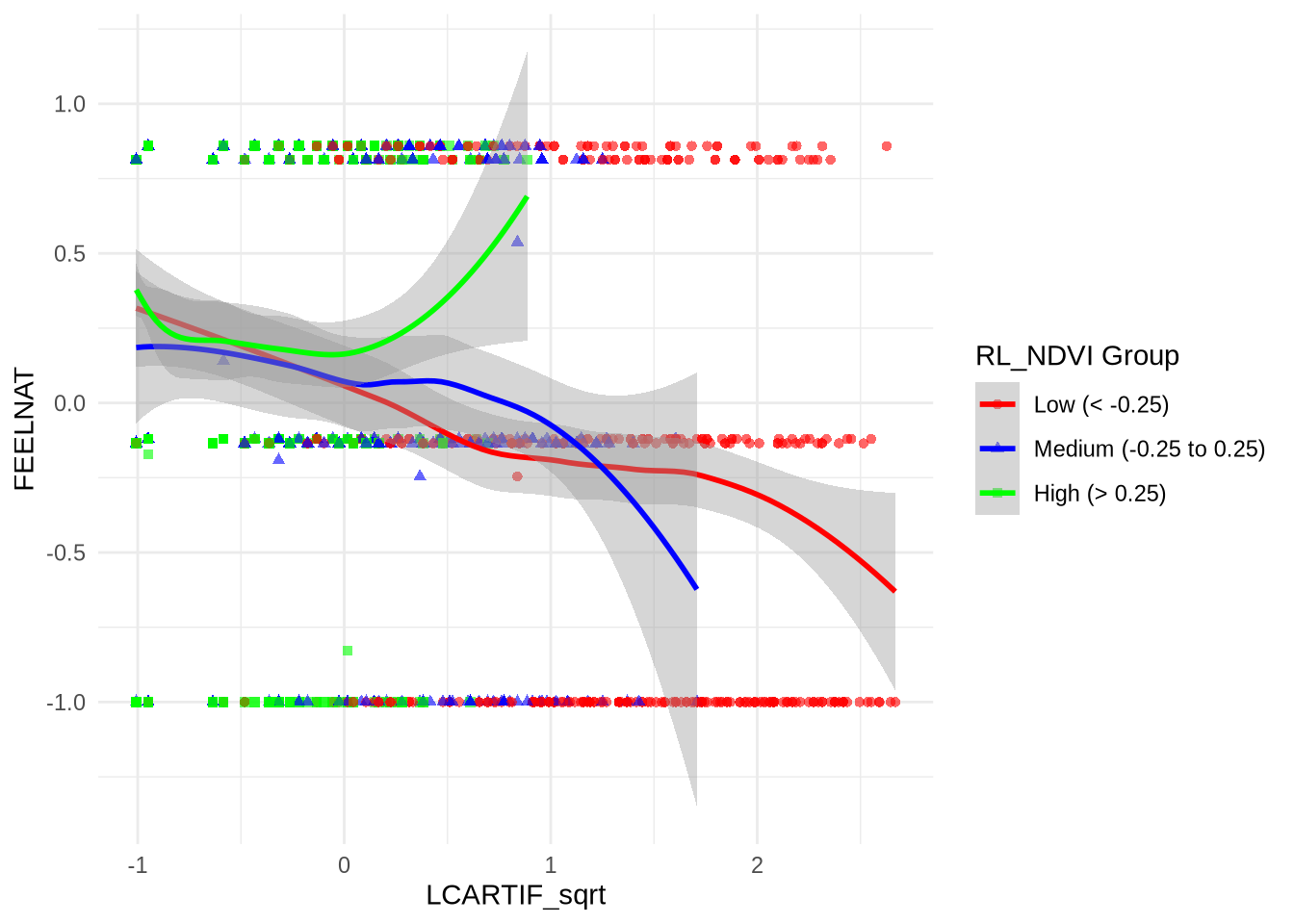
# Elegant function to create coefficient tables from model summaries
suppressPackageStartupMessages({
library(dplyr)
library(tidyr)
library(knitr)
library(purrr)
})
#' Create a formatted coefficient table from model summary list
#'
#' @param model_summaries List of model summaries (e.g., output from lapply(models, summary))
#' @param sig_threshold Significance threshold for bold formatting (default: 0.001)
#' @param covariate_order Optional vector specifying order of covariates
#' @return Formatted table with models as columns and covariates as rows
create_coef_table <- function(model_summaries, sig_threshold = 0.001, covariate_order = NULL) {
# Extract and format coefficients for all models
format_model_coef <- function(coef_matrix, model_name) {
estimates <- coef_matrix[, "Estimate"]
p_values <- coef_matrix[, "Pr(>|t|)"]
# Format with significance stars (common notation)
formatted_coef <- sapply(seq_along(estimates), function(i) {
est_str <- sprintf("%.3f", estimates[i])
stars <- case_when(
p_values[i] < 0.001 ~ "***",
p_values[i] < 0.01 ~ "**",
p_values[i] < 0.05 ~ "*",
p_values[i] < 0.1 ~ ".",
TRUE ~ ""
)
paste0(est_str, stars)
})
tibble(
Model = model_name,
Covariate = rownames(coef_matrix),
Coefficient = formatted_coef
)
}
# Process all models and create wide table
coef_list <- map(model_summaries, coef)
results_table <- map2_dfr(coef_list, names(coef_list), format_model_coef) %>%
pivot_wider(names_from = Model, values_from = Coefficient, values_fill = "")
# Apply covariate ordering
if (is.null(covariate_order)) {
# Default: Intercept first, then alphabetical
all_covariates <- unique(results_table$Covariate)
covariate_order <- c("(Intercept)", sort(all_covariates[all_covariates != "(Intercept)"]))
}
# Filter and reorder covariates
results_table <- results_table %>%
filter(Covariate %in% covariate_order) %>%
slice(match(covariate_order, Covariate))
kable(results_table,
format = "pipe",
align = c("l", rep("c", ncol(results_table) - 1)))
}